General user manual for 'duflor.gui'
Source:vignettes/general-user-manual.Rmd
general-user-manual.RmdThe GUI provided by this package is a front-end to the duflor-package. It follows that the analysis-functions used under the hood will not be documented in this package’s documentation.
General Structure
In figure 1, the general overview of the GUI can be seen.
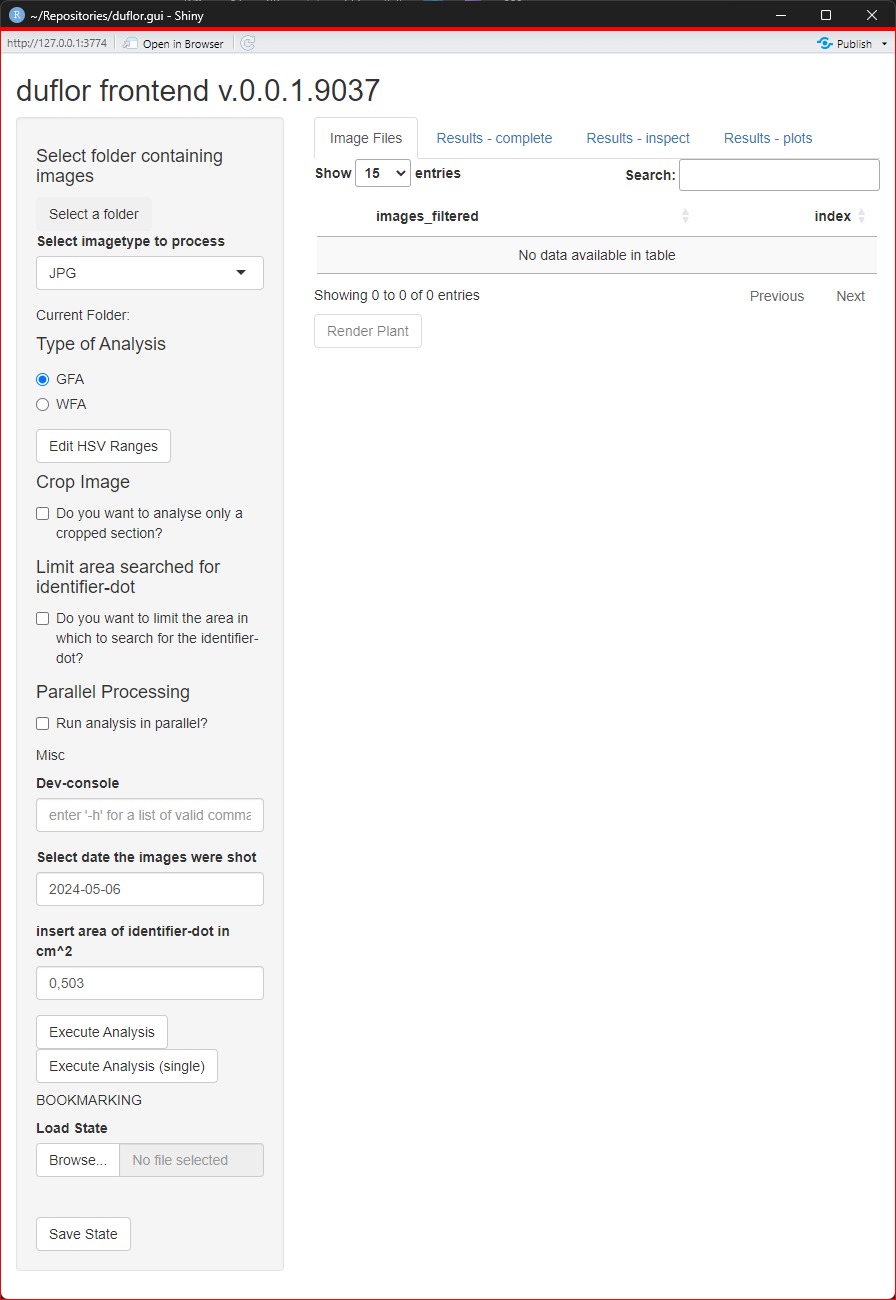
In general, the GUI is split into two regions. The grey panel on the left side contains all input-controls which are relevant for the setup and configuration of an analysis. That is, all steps which must be done to get the results are located here.
Consequently, all buttons/drop-down-menus/tables/other controls which are on the right side of the app are pertaining to the results. It should be noted that several of these controls won’t do anything until the results have been computed1.
Steps required to run an analysis / Input Controls
This section will deal in detail with the controls in the grey panel.
Generally, the steps are designed to be done in order, from top to bottom2:
- Selecting the folder containing the input images.
Note: This folder must not contain any images which are not intended to be evaluated, and this extends to sub-folders. If you select folderfooto be analysed, then imagefoo/bar.pngwill be evaluated. However, the imagesfoo/baz/bar.pngandfoo/baz/another/ridiculous/nesting/plant.pngwill also be evaluated. - Selecting the type of analysis to be performed3. Select
GFAfor aLeaf Area Analysis/Grünflächenanalyse, andWFAif you intend to perform aRoot Area Analysis/Wurzelflächenanalyse. - Optionally, temporarily4 modify the HSV-boundaries used for a
respective type of analysis. Values must be provided in HSV-color space
and in accordance to the following rules:
- , ,
- , ,
-
,
,
Experience suggests that especially the -parameter should be adjusted very carefully. This step is explained in further detail in “Fine-tuning HSV-boundaries”.
- Decide whether or not the image should be cropped. Cropping is explained in more detail in “Cropping Images”.
- Decide whether or not to restrict the area within which the identifier-dot truely lies. For more information, see “Restricting area within which the identifier-dot is quantified”. Taking this precaution is heavily suggested.
- Decide whether or not the execution should be run in parallel. Parallelisation can significantly reduce execution-time, particularly for larger image-sets. It’s use is strongly advised when the number of images gets above ~40. Below that, it might still be sensible, but the author is not comfortable giving a blanket recommendation. For more information, see “Parallelised execution”.
- Select the date at which the images were shot.
- Optionally alter the area for the identifier.
- Decide whether or not to analyse all images listed in the tab
Image Files, or a single one of that set. Pressing either of these buttons will render the GUI unresponsive while executing the respective workflow, until it has been completed and the various results on the right side of the app are being updated.
Post-Execution Steps
After the analysis has finished, the controls of tabs 2-4 on the right side of the app will be updated.
Tab 2: Results - complete
The table in this tab renders the results which will be written to
file
.
At the bottom, you may decide whether or not the results will be saved
as a .csv-file, or as a .xlsx-file. Generally
the use of .csv is suggested for its smaller file-size; but
sometimes the output data must be processed afterwards, and then saving
the data as an .xlsx-file may be advantageous.
Tab 3: Results - inspect
The third tab lets you inspect the results based on a specific spectrum. This table exists for two simple reasons:
- The “complete” table is frequently too crowded to be easily digestible, particularly on lower-DPI screens.
- By selecting a spectrum and a file, the results of applying said spectrum to the selected file can be viewed (see Figure 2). Optionally, the contrast between “hits” and other pixels can be increased. This can be particularly helpful when trying to identify false positive pixels for a given spectrum (see figure 3)
Edge-cases / special controls
State-Management: Saving and restoring application configuration to and from files
At the bottom of the grey panel exists a section called “Bookmarking”. It allows the user to store the current state of the app’s input data (configuration, chosen directory-path, relevant data) to a file, which can be loaded at a later time.
In general, this has two use-cases:
- Retaining and restoring common configuration changes for easier access.
- Restoring configurations which induced errors, so that they might be
debugged in a reproducible manner.
For more details, see
vignette("saving-and-restoring-application-states", package = "duflor.gui").
Tab 1: Image Files
This tab contains the image files which will be analysed. However, it also allows you to view an image. This is particularly useful for verifying that you correctly cropped the image, and/or correctly selected the area for the identifier-dot (if you did any of these suggested extra steps).
An example of how the application of both would look like can be seen in figure 4.
Measures for optimising execution speed
Cropping Images
A very effective way of increasing execution time is to reduce the size of the data- set which is being analysed.
After the desired area has been selected as described above, it can
be optionally reviewed on the same (or another) image by pressing the
Render plant-button on the tab
Image Files.
An in-depth description can be found in
vignette("image-cropping", package = "duflor.gui").
Parallelised execution
Based on the power of the system running the app, one might consider parallelising the evaluation of images to further reduce computation time. However, its effectiveness depends heavily on the resources available.
An in-depth description can be found in
vignette("parallelisation", package = "duflor.gui").
Measures for maximising accuracy
Restricting area within which the identifier-dot is quantified
Typically, the largest error-source has been a bad or inconsistent
determination of the identifier used to calculate the area of extracted
spectra. For information on how to counter this issue, an in-depth
description can be found in
vignette("identifier-cropping", package = "duflor.gui").
Fine-tuning HSV-boundaries
Determining the HSV-boundaries can be an arduous and time-consuming
process. It is not recommended to frequently do this via the
GUI, but instead to create a small script calling the relevant functions
of the duflor-package itself.
An in-depth guide and further details can be found in
vignette("modifying-hsv-bounds", package = "duflor.gui").
(Advanced) Debugging
Generally, the app tries to give reasonable warnings and
status-messages via small red/yellow/blue message boxes. These should be
fairly self-explanatory, and will pop up where the author thought them
sensible or necessary.
If the app runs into an error, in most cases the configuration required
for triggering said error is preserved. For further info, see the
section “Saving states when errors occur during analysis” in
vignette("saving-and-restoring-application-states", package = "duflor.gui").
Development, Modifications, Issues
For details on how to raise new feature-requests, please refer to
vignette("raising-issues", package = "duflor.gui").
Reporting issues
This app, and its underlying code can be reviewed with minimal
upfront work by opening the source-code from GitHub - either by manually
downloading it, or by cloning it via git. To do either,
navigate to the duflor- or duflor.gui-repository.
For details on doing so see
vignette("raising-issues", package = "duflor.gui").